Quick start#
Installation#
The Python module can be directly installed from PyPI with:
pip install anta_database
This Python module is designed to query and visualized data in a AntADatabase folder. The latter has to be first compiled using published IRH datasets, or you can contact for downloading it. You can already have a look at this guide to have an idea of the features of this tool. This Jupyter Notebook can be directly downloaded (top bar) and ran locally (assuming you had downloaded or compiled the AntADatabase folder).
Browsing the database#
First, initialize the Database class by providing the full path to the AntADatabase:
from anta_database import Database
db = Database('/home/anthe/documents/data/isochrones/AntADatabase/')
Use the query() function to browse the database. ‘query()’ without argument will return all metadata contained in the database:
db.query()
Metadata from query:
- author: Franke_2025, Beem_2021, Chung_2023, Cavitte_2020, Mulvaney_2023, Wang_2023, Sanderson_2024, Winter_2018
- age: 4700, 4800, 7200, 10000, 10516, 10700, 11100, 15400, 16800, 25600, 29100, 29300, 30397, 31400, 36300, 37600, 38000, 38100, 39215, 46600, 47110, 48000, 51400, 52200, 62235, 72500, 73100, 73300, 73400, 74000, 75160, 75300, 80007, 81900, 82646, 84300, 90000, 91000, 93900, 95500, 96500, 97400, 97667, 108315, 108600, 113000, 121000, 122800, 128542, 128800, 132000, 135600, 159600, 160000, 161000, 162000, 165200, 166112, 169100, 179000, 199700, 202334, 203000, 215000, 219500, 239000, 240397, 243000, 253400, 262159, 278000, 304000, 320000, 326168, 326800, 336000, 340400, 345462, 365000, 367800, 381523, 395802, 396000, 400000, 414644, 420000, 473000, 483000, 516000, 559000, 610000, 705000
- var: BasalUnit, BedElev, IceThk, SurfElev
- trace_id: 20231211_02_011, VCD_JKB2g_DVD01a, 20231211_01_011, MCM_JKB1a_WLKX01a, 19973310, BV46B-E_proc2, SPC_GCX0e_Y89a, BV34B-BV34E_proc2, IRH_20172047, BV25E-RAID2_proc2, OIA_JKB2n_Y82a, BH07B-BH07E_proc2, HRA3-HRA4_proc2, 20231211_02_012, 20181227_02_005, 20181226_02_009, PNV13___, P09A_PPVV_L4_NS1536_SKIP78_DBS5HZ, 20023111, 19972308, BH45B-BH45E_proc, IRH_20172032, 20181227_02_008, AV01E-RAID1_proc2, AV01B-AV01E_proc2, 20231211_02_014, 20023120, IRH_20172031, 20181226_01_012, 20181226_01_004, RAID2-BV11E_proc2, BV15E-BV15B5_proc2, EPICA_IV_15, BH28E-BH28BB_proc2, PNV07ETOB, DoCo_2007_08_08, 20181222_01_005, 10BDLRSAR_9MI_10, HRA5-HRA6_proc2, 19972206, VCD_JKB2g_X11a, PNH11B-PNH11E_proc2, PNV17___, 20181227_02_001, DoCo_2007_08_13, BV36E-B_proc2, 20231211_02_008, ASB_JKB1a_R11Ea, RAID2-BV28B_proc2, HRB2-HRB3_proc2, BH45E-RAID2_proc, BV11B6-RAID2_proc2, 19983403, IRH_20172046, PNH13E-PNH13B_proc2, 20231211_02_007, 20231211_02_013, OIA_JKB2n_Y88a, 19972405, HRA3-HRA5_proc2, 19983256, EPICA_IV_12, PNH5E-PNH5B_proc2, BH09E-BH09B_proc2, IRH_20172040, 19993135, 20231211_02_009, 20231211_02_017, RAID2-BH35B_proc2, IRH_20172030, SPC_GCX0d_X115a, ASB_JKB1a_R09Ea, PNH15B-DC_proc2, 20231211_01_008, 20231211_02_002, 20181226_01_001, 20231129_01_004, 20231211_01_017, RAID2-BH01E_proc2, PNV03ETOB, LDC-VHF_20201010, BH39B-BH39E_proc2, 20231129_01_015, LDC-VHF_20201012, BH03BB-BH03E_proc2, 19983206, 19983416, DC_PNV09B_proc2, LDC-VHF_20201004, SPC_GCX0e_X114a, 2H30E-H30B, 19983205, IRH_20172029, 19993137, AV02E-AV02B_proc2, A65B_DLRSAR_9MI, WSB_JKB1a_R36a, 19983208, RAID2-DCR210-BH16B_proc2, 19993131, BV38B-E_proc2, 20231211_01_020, 20033138, PNH15ETOB, EPICA_VI_34, OIA_JKB2n_Y77a, A63_DLRSAR_9MI, PNH3B-PNH3E_proc2, 20181226_01_008, BV19E-BV19B5_proc2, SPC_GCX0e_Y93a, 20022116, 19993136, ASB_JKB1a_R11Wa, 19972307, PNV05BTOE, IRH_20172045, PNH7B-PNH7E_proc2, 19972415, OIA_JKB2n_Y68a, OIA_JKB2n_X66a, 20231211_01_019, HRA6-RAID2_proc2, HRA7-HRA8_proc2, BH43B-BH43E_proc2, 19993107, BH11B-BH11E_proc2, HRB2-HRB7-2_proc2, IRH_20172027, DC_LDC_DIVIDE, 19983402, 19973210, 19972304, 19972409, 20231129_01_012, BH18B-BH18E_proc2, SPC_GCX0e_Y85a, A10BDLRSAR_9MI_10, ASB_JKB2e_LKC01a, RAID2-BH21B_proc2, VCD_JKB1a_X08a, 20083126, 20013121, A64B_DLRSAR_9MI, 19983417, 20181226_01_003, 20181227_02_007, 20181226_01_006, 20022104, BH26BB-BH26E_proc2, 19993109, AV04E-AV04B_proc2, 20023110, IRH_20172049, EPICA_II_04, 19972404, EPICA_VIII_23, SPC_GCX0e_X120a, BH32E-BH32B10_proc2, BH01E-BH01B_proc2, 19983102, 20231129_01_011, H42B-2H42E_proc2, 19972313, 20231129_01_008, ASB_JKB1a_R09Ta, 20231211_02_019, 20231211_02_015, MCM_JKB1a_EDMC01a, 2V10E-2V11B, 20231211_02_010, EPICA_VII_27, 2H25E-H25B, PNH1B-DC_proc2, ICP7_JKB2n_F16T04a, DMC_JKB1a_ATAL01e, IRH_20172041, DoCo_2007_08_10, WSB_JKB2g_R34a, 19993104, 19993113, 19983204, HRA2-HRA3_proc2, WEGAS_2010_11_33, WSB_JKB2g_R35c, 20231129_01_017, 20033139, 20013012, DC-LDCRAID2_proc2, OIA_JKB2n_X45a, PNH19ETOB, 20231211_02_018, 20053107, OIA_JKB2n_Y64a, HRA5-HRB6-2_proc2, AV03B-AV03E_proc2, EPICA_II_08, 20231129_01_018, 20053109, A65D_DLRSAR_9MI, 20181226_01_011, 20181226_02_006, H42B-2H42E, 20231211_02_005, EPICA_II_10, SOUTH_PATCH_FLOW_LINE_proc2, 20023151, 20231129_01_010, OIA_JKB2n_X39a, 20181226_01_007, LDC-DC-PANO_proc2, A06CDLRSAR_9MI_clipped, EPICA_VII_26, 20013123, 20173003, BV26E-RAID2_proc2, A64_DLRSAR_9MI, ASB_JKB1a_R10Wa, 20113119, DoCo_2007_08_07, 20181227_03_007, 20231211_01_009, OIA_JKB2n_Y84a, BH24B10-RAID2_proc2, OIA_JKB2n_Y72a, LINK_ROUTE_proc2, BH24E-BH24B10_proc2, ASB_JKB2h_GL0249a, RAID2-BV48E_proc2, IRH_20172025, 11ADLRSAR_9MI_11, A68B_DLRSAR_9MI, 20013010, IRH_20172048, 20181226_01_010, OIA_JKB2n_X57a, 19993120, IRH_20172052, 20013011, OIA_JKB2n_Y80a, A11ADLRSAR_9MI_11, 20231129_01_013, 20023148, OIA_JKB2n_Y75a, OIA_JKB2n_Y60a, 20231211_01_022, BH30BB-BH30E_proc2, SPC_GCX0d_Y83a, OIA_JKB2n_Y81a, RSAR_9MI_clipped, 20231211_01_016, BH41E-BH41B_proc2, AGAP_South_track, BH17M-BH17E_proc2, BH47E-BH47B_proc, PNV19___, 20181226_03_004, 20023149, EPICA_IV_22, BV32B-BV32E_proc2, BH05E-BH05B_proc2, IRH_20172038, 20181227_03_004, 20181226_02_008, 19993115, 20032137, 20181226_01_002, 19972406, 20231129_01_006, 20181226_01_009, 20023153, 19993134, ICP7_JKB2n_F17T01a, BH31E-BH31B_proc, OIA_JKB2n_Y86a, BH15E-BH15M_proc2, WSB_JKB1a_R38a, IRH_20172039, 19993132, ASB_JKB1a_Y38a, IRH_20172026, 19983418, 19993106, BV48E-BV48B_proc2, 20231129_01_016, PNV15___, 20231211_02_006, ICP7_JKB2n_RIDGE1a, 19983401, 19983101, 20231211_02_021, BH36E-BH36B_proc2, HRB6-HRB7_proc2, 20231211_01_010, 20013124, EPICA_VIII_22, 20181227_02_002, SPC_GCX0d_Y75a, HRB1-HRB2, 19983407, LDC-VHF_20201005, 20231204_01_010, OIA_JKB2n_X72a, ASB_JKB1a_R11Ta, 20023105, A08DLRSAR_9MI_3, 19993112, 20181226_03_003, 19983404, 20181227_02_010, EPICA_VII_25, HRB7-SUBCAMP_proc2, CAT-ROUTE-DC_proc2, 20181227_03_002, 20181227_02_009, IRH_20172028, OIA_JKB2n_Y90a, A64E_DLRSAR_9MI, 20181226_02_004, OIA_JKB2n_Y52a, 19972402, BV27E-BV27B_proc2, HRB8-RAID2, 19983406, BH13E-BH13B_proc2, WSB_JKB2h_R40b, 20231211_02_001, 20181227_03_005, ASB_JKB1a_R09Wa, SPC_GCX0d_X112a, 20181226_03_002, SPC_GCX0d_Y79a, VCD_JKB2g_X13a, PNH9E-PNH9B_proc2, 19972303, EPICA_IV_21, 20023152, 19993118, 19983253, BH01B-BH01BB_Proc2, ICP4_JKB2g_F19T03a, 2V02E-RAID, IRH_20172043, A09A_DLRSAR_9MI, 20231211_01_018, 20181227_02_011, PNH17B-PNH17E_proc2, 20173002, 20023109, SPC_GCX0d_Y87a, 20023115, 20023119, 19993026, A07C_DLRSAR_9MI, BV30E-BV30B_proc2, IRH_20172044, 20181226_01_005, RAID2-BH32E_proc2, ICP7_JKB2n_F12T01b, 20023150, HRA1-HRA2_proc2, A67_DLRSAR_9MI, LDC-VHF_20201003, 20181227_02_012, 20181222_01_006, 20181222_01_002, PNV11___, 20231129_01_014, LDC-VHF_20201001, EPICA_III_07, BH21B-BH21E_proc2, 20181227_02_004, WEST_PATCH_FLOW_LINE_proc2, 20231129_01_007, BV44E-BV44B_proc2, 20181226_02_002, BV17B5-BV17E_proc2, 19983457, 19993033, BH39B-BH39E, PNH17BTOE, SPC_GCX0e_X122a, 36_SKIP78_DBS5HZ, 20181226_02_010, 19972309, EPICA_IV_24, 19983207, BV40E-BV40B_proc2, VCD_JKB1a_X09a, 20022106, 20033141, OIA_JKB2n_Y76a, SPC_GCX0e_SPICE1a, 19983408, PNH21BTOE, 20023102, V03E-2V03B, EPICA_VI_35, OIA_JKB2n_X60a, HRB5-HRB2-2_proc2, HRB8-RAID2_proc2, PNV21B__, 20231211_02_020, OIA_JKB2n_X69a, 20231129_01_005, 20231211_01_023, HRA1-HRA2, 20231203_04_006, 19993105, ROUTE5_proc2, 20231211_01_021, HRB1-HRB2_proc2, 20022112, IRH_20172051, 19993127, 20231211_02_003, 20231129_01_020, OIA_JKB2n_X54a, RAID2-BH01B_proc2, 19983254, BV25B5-BV25E_proc2, 20013122, 20131127_01, BV26B-BV26E_proc2, HRB4-HRB5_proc2, LDC-VHF_20201007, HRB6-HRB3-2_proc2, EPICA_III_18, BH42E-BH42B_proc2, OIA_JKB2n_X63a, A07B_DLRSAR_9MI, 20231211_01_013, ASB_JKB1a_R08Ec, LDC-VHF_20201002, 19993116, 20181222_01_001, 20181222_01_004, BV23E-BV23B5_proc2, BH37E-BH37B_proc2, EPICA_IV_23, 20181227_03_001, BV50B-BV50E_proc, DoCo_2007_08_12, BH33B-BH33E_proc, HRA4-HRA6_proc2, 19972408, 2V23B-2V23E, 20231211_02_004, OIA_JKB2n_Y15a, HRB7-HRB8, LDC-VHF_20201011, 20231129_01_009, BH40B-BH40E_proc2, OIA_JKB2n_Y74a, DC_LDCRAID_proc2, HRB7-HRB8_proc2, BH03B-BH03E_proc2, BV13B5-BV13E_proc2, P34A_PS2_L4_NS1536_SKIP78_DBS5HZ_04, SPC_GCX0e_X125a, 20181226_03_001, EPICA_IV_10, 20181227_02_006, A66_DLRSAR_9MI, BV11E-BV11B6_proc2, 20181227_01_001, 20033143, LDC-VHF_20201006, 20023154, PNH1E-PNH1B_proc2, BH03E-RAID2_proc2, OIA_JKB2n_Y70b, P26A_PS2_L4_NS1536_SKIP78_DBS5HZ, BV21B5-BV21E_proc2, LDC-VHF_20201008, P24A_PS2_L4_NS1536_SKIP78_DBS5HZ, OIA_JKB2n_Y78a, 20181227_02_014, 20181227_03_008, RAID2-BV42B_proc2, SPC_GCX0e_Y98a, 20181226_01_013, BH19E-BH19B_proc2, 20022115, 19983202, 20022107, 20181227_02_015, V18B-V18E, BV52E-BV52B_proc, OIA_JKB2n_X48a, RAID2-BH36E_proc2, 19993122, 20173004, RAID-2V11E, BH43E-RAID2_proc2, 20231211_01_006, EPICA_IV_11, 20231211_02_016, LDC-VHF_20201009, OIA_JKB2n_Y25a, BH34E-BH34B10_proc2, BV42B-BV42E_proc2, OIA_JKB2n_Y79a, BH38E-BH38B_proc2, GPS_FLOW_LINE_proc2, 20231211_01_014, 20231211_01_012, 20181227_03_003, BV34B-BV34E-2_proc2, 20231211_01_007, ICP7_JKB2n_EDMC02a, PNV01BTOE, PNV21B-DC_proc2, 20231211_01_024, AV05B-AV05E_proc2, ROUTE4_proc2, VCD_JKB2g_X08b, LDCCAT-SUBCAMP_proc2, 20231211_01_015, 19993110, IRH_20172050, 19983201, BV28B-BV28E_proc2
- reference: Franke et al. (2025), Beem et al. (2021), Chung et al. (2023), Cavitte et al. (2020), Mulvaney et al. (2023), Wang et al. (2023), Sanderson et al. (2024), Winter et al. (2018)
- DOIs: https://doi.org/10.1594/PANGAEA.973266, https://doi.org/10.15784/601437, https://doi.pangaea.de/10.1594/PANGAEA.957176, https://doi.org/10.15784/601411, https://doi.pangaea.de/10.1594/PANGAEA.963470, https://doi.org/10.1594/PANGAEA.958462, https://doi.org/10.5285/cfafb639-991a-422f-9caa-7793c195d316, https://doi.org/10.1594/PANGAEA.895528
- database: /home/anthe/documents/data/isochrones/AntADatabase//AntADatabase.db
- query params: {'age': None, 'var': None, 'author': None, 'trace_id': None}
The query() function takes as argument:
author: author(s) of the datasets of interest
age: age(s) in yrs before present of the layer(s) of interest
var: variables(s) other than age depth such as ‘IceThk’, ‘SurfElev’, ‘BedElev’ or ‘BasalUnit’
trace_id: ID of a particular trace. This is useful for explicit trace IDs such as ‘DC_LDC_DIVIDE’
One can also combine field queries as well as providing lists. Here are a few examples:
#Examples of queries:
db.query(author='Cavitte_2020') # all data from Cavitte et al. 2020
db.query(age='38100') # all datasets with the 38.1ka isochrone
db.query(var='IceThk') # all datasets with IceThk variable
db.query(trace_id='DC_LDC_DIVIDE') # all layers with the trace ID DC_LDC_DIVIDE
db.query(author=['Franke_2025', 'Winter_2018'], age='38100') # example of multiple criteria
Metadata from query:
- author: Franke_2025, Winter_2018
- age: 38100
- var:
- trace_id: 20181227_02_004, 20231129_01_007, 20181226_02_002, 20231211_02_011, 19993033, 20231211_01_011, 19973310, 19983402, 19972304, 20231211_01_016, 20181226_02_010, 19972309, EPICA_IV_24, 19972409, AGAP_South_track, 19983207, 20231129_01_012, 20022106, 20033141, 20083126, 20231211_02_012, 20181226_03_004, 20023149, 20181227_02_005, 20181226_02_009, EPICA_IV_22, 20013121, 19983408, 20023111, 19972308, 20181227_03_004, 20181226_02_008, 19993115, 20181227_02_008, EPICA_VI_35, 20181226_01_003, 20032137, 20181226_01_002, 19972406, 20181227_02_007, 20181226_01_006, 20231129_01_006, 20181226_01_009, 20022104, 20231211_02_014, 20023120, 20023153, 20181226_01_012, 19993134, 20181226_01_004, EPICA_IV_15, 19993109, 20231211_02_020, 20231129_01_005, 20023110, 20231211_01_023, EPICA_II_04, 19972404, 20181222_01_005, DoCo_2007_08_08, 20231203_04_006, EPICA_VIII_23, 19993105, 20231211_01_021, 19993132, 19972206, 20022112, 19983102, 20181227_02_001, 20231129_01_011, 20231211_02_003, 19993127, 19993106, 20231129_01_016, DoCo_2007_08_13, 19972313, 20231211_02_006, 20013122, 19983401, 19983101, 20231211_02_021, 20231211_02_008, 20231211_01_010, 20013124, 20231129_01_008, 20231211_02_019, 20231211_02_015, 19983403, EPICA_VIII_22, EPICA_III_18, 20181227_02_002, 20231211_02_007, 20231211_02_013, 20231211_01_013, EPICA_IV_23, 19993116, 20181222_01_001, 20181222_01_004, 19972405, 20231211_02_010, EPICA_VII_27, 19983407, 20181227_03_001, DoCo_2007_08_12, 20231204_01_010, EPICA_IV_12, 19972408, 19993135, 20231211_02_004, 20231211_02_009, 20231211_02_017, 20023105, DoCo_2007_08_10, 19993112, 20231129_01_009, 20181226_03_003, 19983404, 19993104, 20231211_01_008, 20181227_02_010, 19993113, 20231211_02_002, EPICA_VII_25, 19983204, 20181226_03_001, EPICA_IV_10, WEGAS_2010_11_33, 20181227_03_002, 20181227_02_009, 20181227_02_006, 20181226_01_001, 20181226_02_004, 20013012, 20231129_01_017, 20231129_01_004, 19972402, 20033143, 20231211_01_017, 20231211_02_018, 19983406, 20023154, 20231211_02_001, 20181227_03_005, 20231129_01_015, EPICA_II_08, 19983206, 20181226_03_002, 20231129_01_018, 19983416, 20053109, 20181226_01_011, 20181226_02_006, EPICA_II_10, 20231211_02_005, 20181227_02_014, 19972303, 20181227_03_008, 20023151, EPICA_IV_21, 19983205, 19993137, 20181226_01_013, 20022115, 19983202, 20022107, 20181227_02_015, 20231129_01_010, 20023152, 20181226_01_007, 19993118, EPICA_VII_26, 20013123, 20173003, 19993131, 19993122, 20113119, 20173004, 20231211_01_006, DoCo_2007_08_07, 20181227_03_007, 20231211_01_020, 20231211_01_009, 20231211_01_018, EPICA_IV_11, 20181227_02_011, 20231211_02_016, 20023109, 20033138, 20023115, 20023119, 20231211_01_014, 19993026, 20231211_01_012, 20181227_03_003, 20231211_01_007, EPICA_VI_34, 20013010, 20181226_01_005, 20231211_01_024, 20181226_01_010, 19993120, 20023150, 20181226_01_008, 20181227_02_012, 20013011, 20022116, 19993136, 20181222_01_006, 19972307, 20231211_01_015, 20181222_01_002, 19993110, 20231129_01_014, 20231129_01_013, EPICA_III_07, 20023148, 19972415, 19983201, 20231211_01_019, 20231211_01_022, 19993107
- reference: Franke et al. (2025), Winter et al. (2018)
- DOIs: https://doi.org/10.1594/PANGAEA.973266, https://doi.org/10.1594/PANGAEA.895528
- database: /home/anthe/documents/data/isochrones/AntADatabase//AntADatabase.db
- query params: {'age': '38100', 'var': None, 'author': ['Franke_2025', 'Winter_2018'], 'trace_id': None}
Visualization#
Use the results of the query in the plotting functions:
results = db.query(age=['38100','38000'])
db.plotXY(results,
downscale_factor=1000, # downscale the datasets n times, which makes no visual difference but it is much lighter
title='AntArchitecture 38(.1)ka layer',
xlim=(-500, 2400), ylim=(-2200, 2200), # set the plot extent in km
scale_factor=0.7, # adjust the size of the plot
latex=True, # use latex compilers for plotting if you have them installed on your system
# save='AntA_38ka.pdf', # Uncomment to save the figure, otherwise it we visualize with pyplot
)
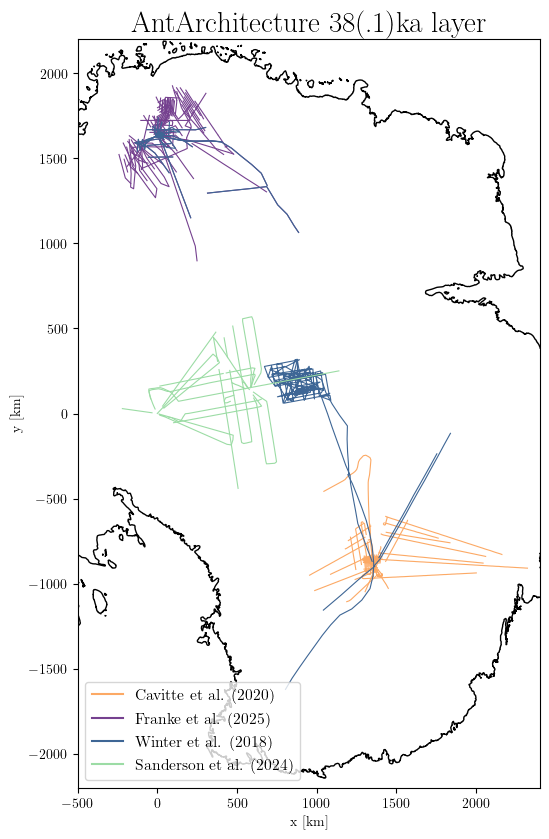
Current implemented plotting functions are:
plotXY(): plot locations of the data, with different colors for the different datasets (example above)
plotXYDepth(): scatter plot with a colormap displaying the values of the data (example below)
In Jupyter Notebook, use ‘%matplotlib qt’ or ‘%matplotlib widget’ depending on your IDE, to switch to the matplotlib widget that allows you to zoom in etc. Use ‘%matplotlib inline’ (default) to plot the figure in the notebook
# %matplotlib widget
%matplotlib inline
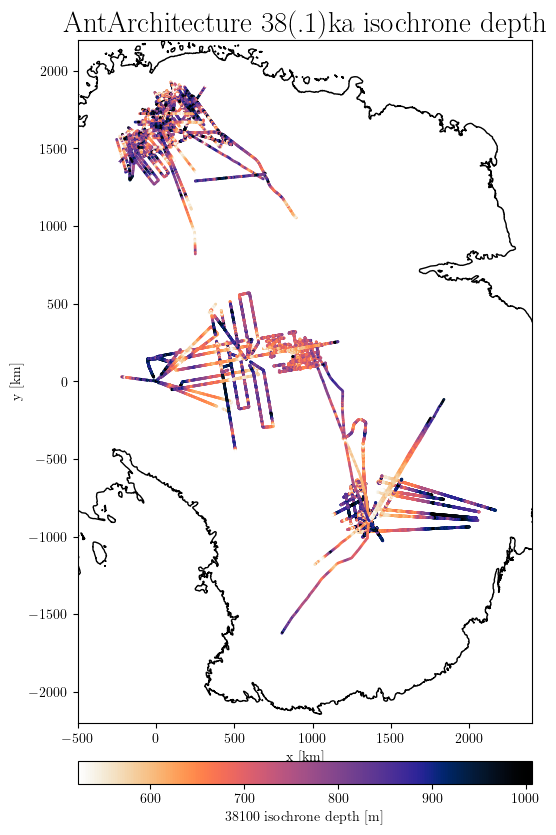
results = db.query(age=['38100','38000'])
db.plotXYDepth(results, title='AntArchitecture 38(.1)ka isochrone depth',
downscale_factor=100,
xlim=(-500, 2400),
ylim=(-2200, 2200),
scale_factor=0.7,
latex=True,
# save='AntA_38ka_depth.png'
)
Generate data from the database#
Note: This part could be developed further in the future if there is the need. But for now, I am designing a separate Python module for constraining my ice sheet model of use, which is tailored to this database and other parallel processing libraries.
The data_generator() function reads the query and ‘yield’ the dataframes for later use. Here is a quick example of how this can be used for computing the mean layer depth:
results = db.query(age=['38100'])
lazy_dfs = db.data_generator()
import numpy as np
mean_depth_trs = []
min_depth = float('inf')
max_depth = float('-inf')
for df, md in lazy_dfs:
depth_values = df[md['age']]
mean_depth_trs.append(np.mean(depth_values))
min_depth = min(min_depth, min(depth_values))
max_depth = max(max_depth, max(depth_values))
mean_depth = np.mean(mean_depth_trs)
std_dev = np.std(mean_depth_trs, ddof=1)
print(f"The mean depth of the 38ka isochrone across East Antarctica is {round(mean_depth, 2)} m ranging from {round(min_depth, 2)} m to {round(max_depth, 2)} m.")
The mean depth of the 38ka isochrone across East Antarctica is 1127.89 m ranging from 313.34 m to 2213.46 m.
In the for loop, ‘df’ is an individual dataframe from the database, corresponding to a single layer of a single trace from a single dataset. The ‘md’ stores the unique metadata from the current ‘df’. One can then associate the metadata with the current df, for example:
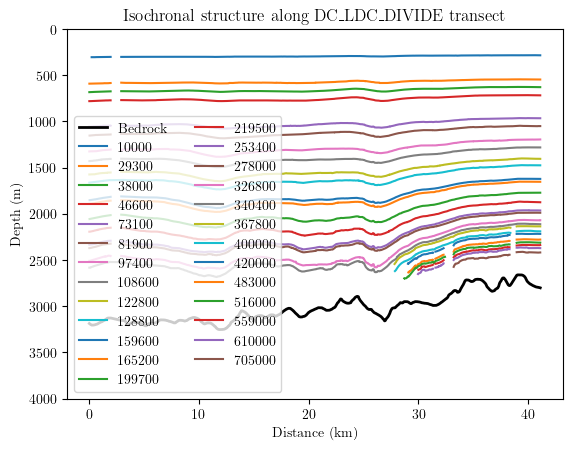
results = db.query(author='Cavitte_2020', trace_id='DC_LDC_DIVIDE')
lazy_dfs = db.data_generator(results)
import matplotlib.pyplot as plt
# from matplotlib import rc
# rc('font', **{'family': 'serif', 'serif': ['Computer Modern']})
# rc('text', usetex=True) # Do you use latex?
for df, md in lazy_dfs:
if md['age'] is not None:
plt.plot(df.distance/1000, df[md['age']], label=md['age'])
if md['var'] == 'IceThk':
plt.plot(df.distance/1000, df['IceThk'], linewidth=2, color='k', label='Bedrock')
plt.legend(ncol=2)
plt.xlabel('Distance (km)')
plt.ylabel('Depth (m)')
plt.ylim(4000, 0)
plt.title('Isochronal structure along DC_LDC_DIVIDE transect')
plt.show()
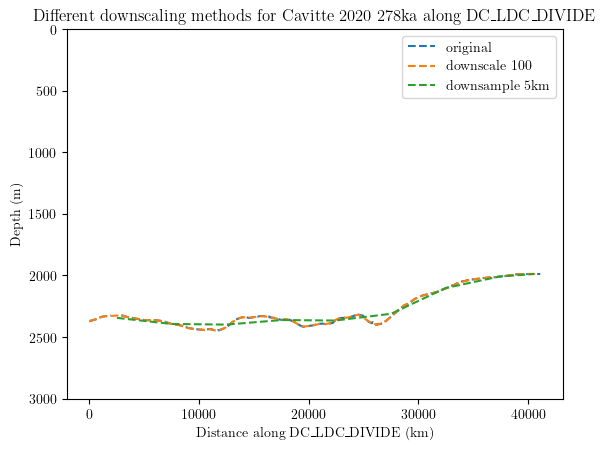
Downscale the data#
The downscale_factor argument in the data_generator reduces the size of the generated data by n times. This is useful when dealing with large data where the downscaling has a neglectable influence on the results. Another solution, probably more straightforward, is the downsample_distance option, which allows to provide a distance in meters by which the data should be average along the transects:
import matplotlib.pyplot as plt
results = db.query(author='Cavitte_2020', age='278000', trace_id='DC_LDC_DIVIDE')
original = db.data_generator(results)
downscale_10 = db.data_generator(results, downscale_factor=100)
downsampled_by_distance = db.data_generator(results, downsample_distance=5000)
list = [original, downscale_10, downsampled_by_distance]
labels = ['original', 'downscale 100', 'downsample 5km']
for i, lazy_dfs in enumerate(list):
for df, md in lazy_dfs:
plt.plot(df.distance, df['278000'],label=labels[i], linestyle='--')
plt.ylim(3000, 0)
plt.xlabel('Distance along DC_LDC_DIVIDE (km)')
plt.ylabel('Depth (m)')
plt.legend()
plt.title('Different downscaling methods for Cavitte 2020 278ka along DC_LDC_DIVIDE')
plt.show()